Announcing ggraph: A grammar of graphics for relational data
February 23, 2017
I am absolutely thrilled to announce that ggraph has finally been released
on CRAN. ggraph is my most ambitious package to date and its very early
genesis has been described in a prior post.
If any mention of ggraph is completely new to you, then in short terms ggraph
is an extension of the ggplot2 API to support relational data such as networks
and trees. I feel fairly confident in saying that ggraph is the most
powerful way to create static network based visualizations in R. Leading up to
the release, the three main concepts of ggraph has been described in detail in
their own blog posts
(layouts,
nodes, and
edges) so this
will not be reiterated here. Instead I’ll talk a bit about the philosophy behind
the package as well as show of some of the features that do not fall into any of
the three main concepts.
The Philosophy
There is no shortage of software for creating network visualizations and there
is no shortage of said visualizations themselves. Often though, the
visualizations are more impressive than informative and it is easy to feel that
their main task is to show that we are really dealing with some complex data.
All of this has led to a certain disdain for classic network visualizations
perfectly encapsulated in the nickname hairballs. It does not have to be like
this! The greatness of ggplot2 lies in how it allows users to quickly iterate
over visualization approaches, thus better ensuring that the best visualization
approach is reached. If this was extended to relational data it is my belief
that users would be more likely to try to make plots that are more meaningful.
After all we all want interpretability, right? Consider having to try out 7
different network visualization packages with different APIs versus just mixing
and matching layouts and geoms in an iterative process — I know which way I
prefer.
The goal of ggraph is thus clear — provide everything related to
visualizations of relational data in a ggplot2-like API to lessen the
cognitive load on experimenting with different visual representations. I’m not
there yet, but I feel the current version represents a solid foundation where
most users will not feel many limitations — on the contrary I believe most
users will feel like the chains have come off and they are set free.
Future focus
As I pointed out, ggraph is far from done. I’ll try to keep my development
focus in the open by putting things on the road-map as
GitHub issues. Honorable mentions
include matrix, d3-force and sankey layout, expanded support for edge endings
(more choices than grid::arrow() provides), edge routing (avoid node
collision), and textbox nodes. I welcome all suggestions as the world of network
visualizations is moving fast and I cannot keep on top of everything.
Features besides layouts, nodes, and edges
Understanding the node and edge geoms along with how layouts are defined will
get you a long way towards visualizing networks. Still, ggraph has more to
offer, some of which will be discussed here:
theme_graph()
Consider the following plot:
library(ggraph)
library(igraph)
graph <- graph_from_data_frame(highschool)
p <- ggraph(graph, layout = 'kk') +
geom_edge_link(aes(colour = factor(year))) +
geom_node_point() +
ggtitle('An example')
p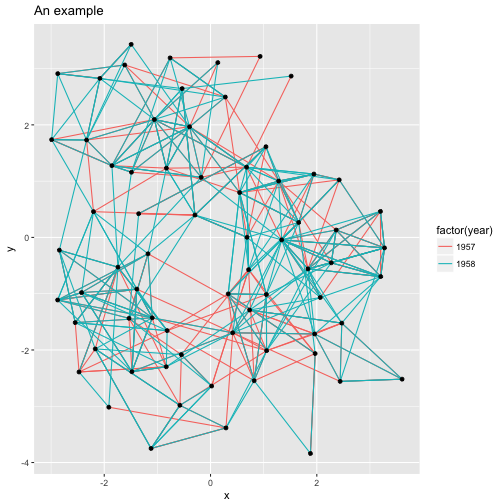
While the ggplot2 heritage clearly shows due to the grey background with white
grid lines, the whole concept of x and y axes is often redundant in network
visualizations and are just a distraction. ggraph provides its own theme
optimized for network visualizations called theme_graph(), that facilitates
clean and beautiful visualizations:
p + theme_graph()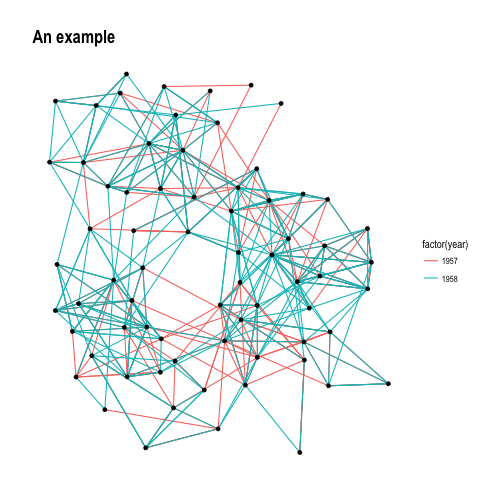
theme_graph(), besides removing axes, grids, and border, changes the font to
Arial Narrow (this can be overridden). Furthermore, it makes it easy to change
the coloring of the plot:
p + theme_graph(background = 'grey20', text_colour = 'white')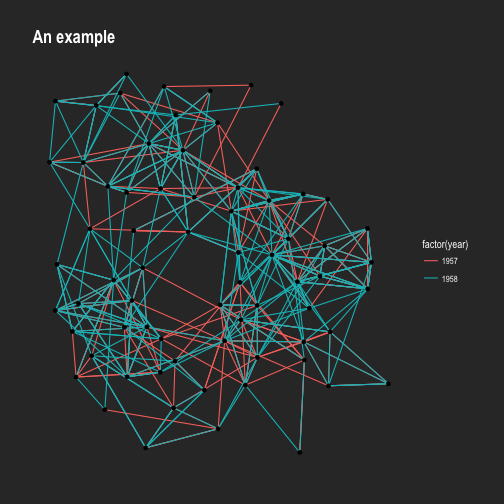
Adding the same theme to every plot is tedious and ggraph provides a way to
avoid this. Using set_graph_style() the theme_graph() is set as default. As
an extra benefit all text-based geoms gets their defaults updated so the text
automatically uses the same style as the theme.
set_graph_style()
p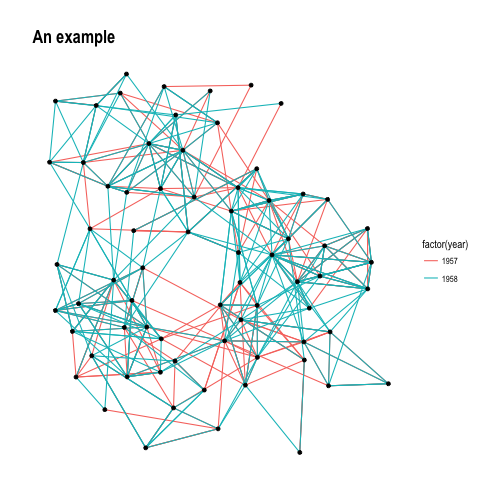
Facetting
A powerful but underutilized way of gaining insight into networks is by using
small multiples. This technique can reduce edge over-plotting in a very
meaningful way by spreading nodes and edges out based on their attributes. The
benefits of small multiples are not unique to relational data, as the popularity
of ggplot2s facetting functionality shows. The base facetting functions
provided by ggplot2 is a bad fit for networks though, as we are working with
two very distinct types of data. If you facet on a node attribute, all edges
would be plotted in all panels, despite the terminal nodes not being present
which is not what you expect. Because of this ggraph comes with its own set
of facetting functions tailored to network data:
facet_nodes() and facet_edges()
These two functions are equivalent to facet_wrap() in functionality, but they
only address node and edge data respectively. When using facet_nodes() edges
are only drawn in a panel if both terminal nodes are present there. When using
facet_edges() nodes are always drawn in all panels even if the node data
contains an attribute named the same as the one used for the edge facetting.
# Assign each node to a random class
V(graph)$class <- sample(letters[1:4], gorder(graph), TRUE)
# Make year a character
E(graph)$year <- as.character(E(graph)$year)
p <- ggraph(graph, layout = 'kk') +
geom_edge_fan(aes(alpha = ..index.., colour = year)) +
geom_node_point(aes(shape = class)) +
scale_edge_alpha(guide = 'none')
p + facet_edges(~year)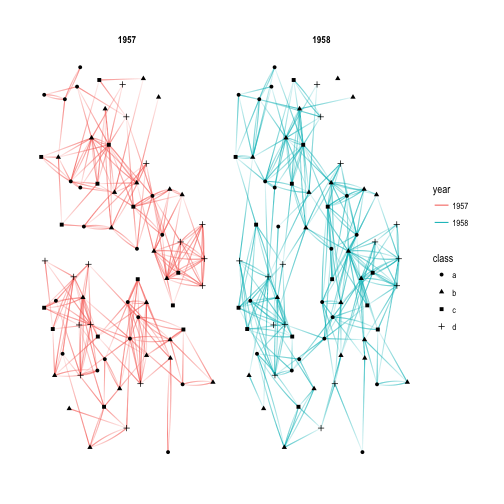
Often, when working with small multiples it is nice to have some visual
separation between each plot — setting a foreground color in theme_graph()
will add strip background and border (you can also use the th_foreground()
helper for this):
p + facet_nodes(~class) + th_foreground(foreground = 'grey80', border = TRUE)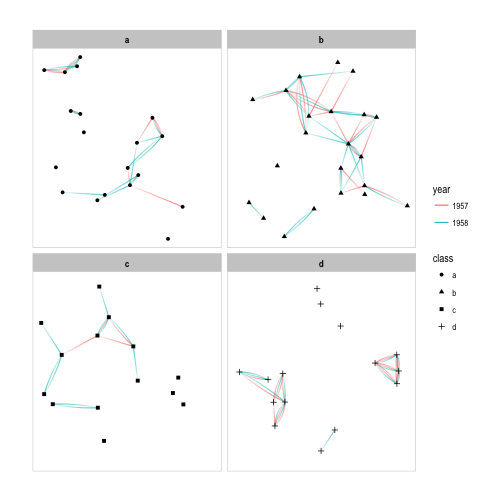
# Lets not have to add this everytime
set_graph_style(foreground = 'grey80')facet_graph
Facetting on two variables simultaneously is very powerful and something that
is supported in ggplot2 with facet_grid(). In ggraph the same is possible
using facet_graph() that takes the behavior of facet_nodes() and
facet_edges() and combines them:
p + facet_graph(year ~ class)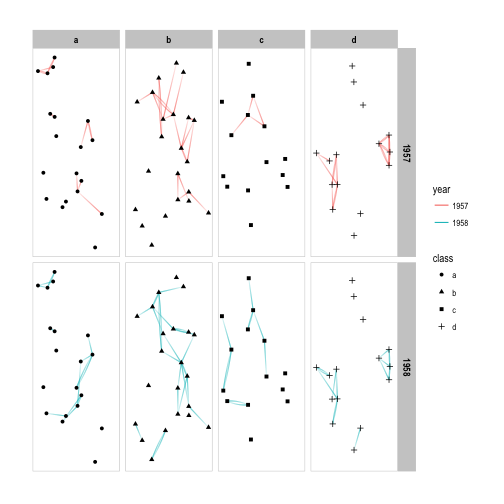
As with facet_grid() marginal plots are supported as well:
p + facet_graph(year ~ class, margins = TRUE)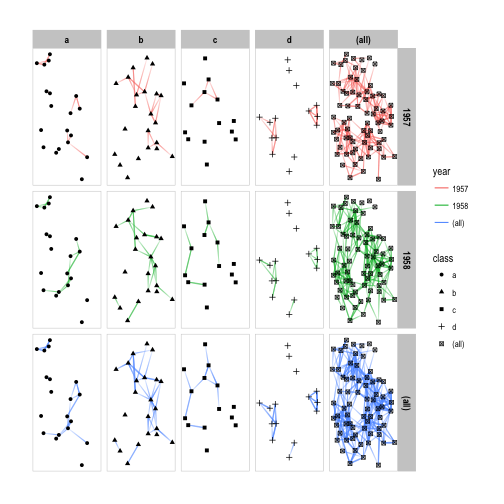
While the default is to put facet the rows on edges and the columns on nodes,
this is free to change using the row_type and col_type arguments. There is
nothing stopping you from facetting on the same type in each dimension either:
# Facet edge by the class of their start node as well as year
p + facet_graph(year ~ node1.class, col_type = 'edge')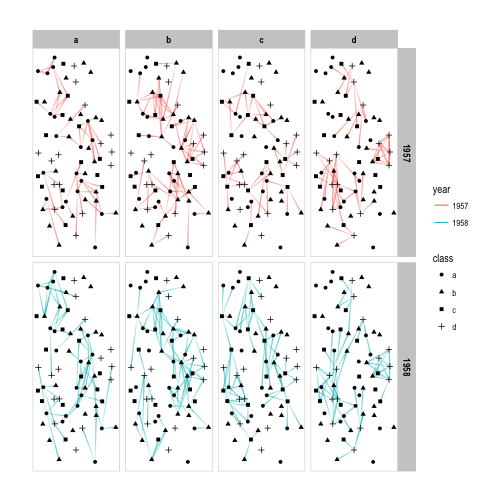
I hope I have convinced you that facetting in the context of relational data is both very easy, as well as extremely powerful. Avoiding the hairball is one of the prime goal of network visualizations and using small multiples is a fantastic way of cutting down on the number of nodes and edges while still getting the full picture.